News
[2024-05-14]
We have currently release our model weights, training code, and tech report. Discussions are welcome.
For training code, please refer to our github
For training details, please refer to our tech-report
[2024-04-22]
piccolo-large-zh-v2 currently ranks first on the C-MTEB list, leading the previous BERT model by about 1.9 points.
Piccolo-large-zh-v2
piccolo-large-zh-v2 is a Chinese embedding model developed by the general model group from SenseTime Research. This upgraded version of Piccolo aims to prioritize general downstream fine-tuning methods. Piccolo2 primarily leverages an efficient multi-task hybrid loss training approach, effectively harnessing textual data and labels from diverse downstream tasks. In addition, Piccolo2 scales up the embedding dimension and uses MRL training to support more flexible vector dimensions.
💡 Model Hightlights
The main feature of piccolo2 is that it uses a multi-task hybrid loss during training.
For retrieval/sorting tasks, we use the standard InfoNCE with in-batch-negative:
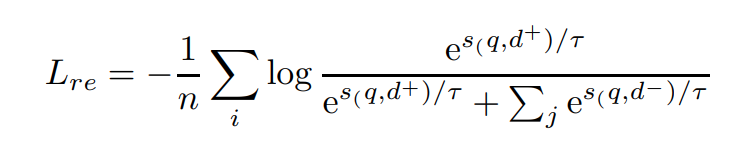
For sts/pair classification tasks, we use cosent loss, which is proved to be better for data with more fine-grained labels(e.g. score values ):
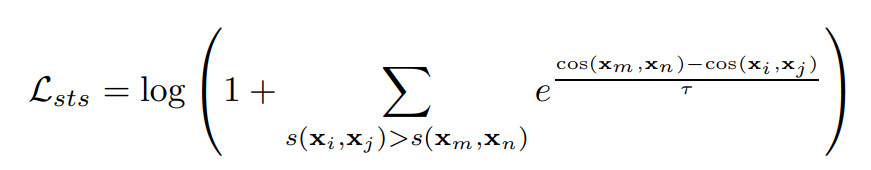
For classification/clustering tasks, by treating text and its semantic labels as positive and negative pairs, we convert the dataset into the format of triples. And then we use InfoNCE to optimize it. However, it’s important to stress that in-batch negatives are no longer used due to the fact that it can easily lead to conflict training targets:
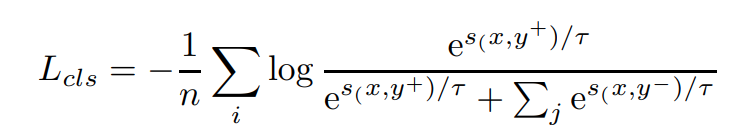
📃 Experiments and Results
Piccolo2 primarily focuses on the downstream general finetune paradigm. Our open source model uses stella-v3.5 as initialization and trained about 2500 steps on 32 GPUS. For more implementation details, please refer to our technical report.
| Model Name | Model Size (GB) | Dimension | Sequence Length | Classification (9) | Clustering (4) | Pair Classification (2) | Reranking (4) | Retrieval (8) | STS (8) | Average (35) |
|---|---|---|---|---|---|---|---|---|---|---|
| piccolo-large-zh-v2 | 1.21 | 1792 | 512 | 74.59 | 62.17 | 90.24 | 70 | 74.36 | 63.5 | 70.95 |
| gte-Qwen1.5-7B-instruct | 26.45 | 32768 | 4096 | 73.35 | 67.08 | 88.52 | 66.38 | 70.62 | 62.32 | 69.56 |
| acge-text-embedding | 1.21 | 1792 | 512 | 72.75 | 58.7 | 87.84 | 67.98 | 72.93 | 62.09 | 69.07 |
🔨 Usage
The piccolo model can be easily accessed in the sentence-transformer package:
# for s2s/s2p dataset, you can use piccolo as below
from sklearn.preprocessing import normalize
from sentence_transformers import SentenceTransformer
sentences = ["数据1", "数据2"]
matryoshka_dim=1792 # support 256, 512, 768, 1024, 1280, 1536, 1792
model = SentenceTransformer('sensenova/piccolo-large-zh-v2')
embeddings_1 = model.encode(sentences, normalize_embeddings=False)
embeddings_2 = model.encode(sentences, normalize_embeddings=False)
embeddings_1 = normalize(embeddings_1[..., :matryoshka_dim], norm="l2", axis=1)
embeddings_2 = normalize(embeddings_2[..., :matryoshka_dim], norm="l2", axis=1)
similarity = embeddings_1 @ embeddings_2.T
🤗 Model List
| Model | Language | Description | prompt |
|---|---|---|---|
| sensenova/piccolo-large-zh-v2 | Chinese | version2: finetuning with multi-task hybrid loss training | None |
| sensenova/piccolo-large-zh | Chinese | version1: pretrain under 400 million chinese text pair | '查询'/'结果' |
| sensenova/piccolo-base-zh | Chinese | version1: pretrain under 400 million chinese text pair | '查询'/'结果' |
Citation
If you find our tech report, models or code helpful, please cite our report or give a star on github or huggingface!
@misc{2405.06932,
Author = {Junqin Huang and Zhongjie Hu and Zihao Jing and Mengya Gao and Yichao Wu},
Title = {Piccolo2: General Text Embedding with Multi-task Hybrid Loss Training},
Year = {2024},
Eprint = {arXiv:2405.06932},
}
- Downloads last month
- 5
Evaluation results
- cos_sim_pearson on MTEB AFQMCvalidation set self-reported56.761
- cos_sim_spearman on MTEB AFQMCvalidation set self-reported61.493
- euclidean_pearson on MTEB AFQMCvalidation set self-reported59.145
- euclidean_spearman on MTEB AFQMCvalidation set self-reported60.636
- manhattan_pearson on MTEB AFQMCvalidation set self-reported59.147
- manhattan_spearman on MTEB AFQMCvalidation set self-reported60.635
- cos_sim_pearson on MTEB ATECtest set self-reported56.217
- cos_sim_spearman on MTEB ATECtest set self-reported59.198
- euclidean_pearson on MTEB ATECtest set self-reported62.378
- euclidean_spearman on MTEB ATECtest set self-reported58.794
- manhattan_pearson on MTEB ATECtest set self-reported62.370
- manhattan_spearman on MTEB ATECtest set self-reported58.792
- accuracy on MTEB AmazonReviewsClassification (zh)test set self-reported49.440
- f1 on MTEB AmazonReviewsClassification (zh)test set self-reported46.674
- cos_sim_pearson on MTEB BQtest set self-reported70.990
- cos_sim_spearman on MTEB BQtest set self-reported72.876
- euclidean_pearson on MTEB BQtest set self-reported71.177
- euclidean_spearman on MTEB BQtest set self-reported72.504
- manhattan_pearson on MTEB BQtest set self-reported71.173
- manhattan_spearman on MTEB BQtest set self-reported72.498
- v_measure on MTEB CLSClusteringP2Ptest set self-reported57.927
- v_measure on MTEB CLSClusteringS2Stest set self-reported48.097
- map on MTEB CMedQAv1test set self-reported89.310
- mrr on MTEB CMedQAv1test set self-reported91.381
- map on MTEB CMedQAv2test set self-reported90.138
- mrr on MTEB CMedQAv2test set self-reported92.143
- map_at_1 on MTEB CmedqaRetrievalself-reported26.931
- map_at_10 on MTEB CmedqaRetrievalself-reported40.647
- map_at_100 on MTEB CmedqaRetrievalself-reported42.519
- map_at_1000 on MTEB CmedqaRetrievalself-reported42.616
- map_at_3 on MTEB CmedqaRetrievalself-reported36.145
- map_at_5 on MTEB CmedqaRetrievalself-reported38.717
- mrr_at_1 on MTEB CmedqaRetrievalself-reported40.935
- mrr_at_10 on MTEB CmedqaRetrievalself-reported49.684
- mrr_at_100 on MTEB CmedqaRetrievalself-reported50.598
- mrr_at_1000 on MTEB CmedqaRetrievalself-reported50.633
- mrr_at_3 on MTEB CmedqaRetrievalself-reported47.070
- mrr_at_5 on MTEB CmedqaRetrievalself-reported48.490
- ndcg_at_1 on MTEB CmedqaRetrievalself-reported40.935
- ndcg_at_10 on MTEB CmedqaRetrievalself-reported47.584
- ndcg_at_100 on MTEB CmedqaRetrievalself-reported54.692
- ndcg_at_1000 on MTEB CmedqaRetrievalself-reported56.314
- ndcg_at_3 on MTEB CmedqaRetrievalself-reported41.973
- ndcg_at_5 on MTEB CmedqaRetrievalself-reported44.334
- precision_at_1 on MTEB CmedqaRetrievalself-reported40.935
- precision_at_10 on MTEB CmedqaRetrievalself-reported10.585
- precision_at_100 on MTEB CmedqaRetrievalself-reported1.637
- precision_at_1000 on MTEB CmedqaRetrievalself-reported0.184
- precision_at_3 on MTEB CmedqaRetrievalself-reported23.881
- precision_at_5 on MTEB CmedqaRetrievalself-reported17.399
- recall_at_1 on MTEB CmedqaRetrievalself-reported26.931
- recall_at_10 on MTEB CmedqaRetrievalself-reported59.006
- recall_at_100 on MTEB CmedqaRetrievalself-reported88.247
- recall_at_1000 on MTEB CmedqaRetrievalself-reported99.045
- recall_at_3 on MTEB CmedqaRetrievalself-reported42.064
- recall_at_5 on MTEB CmedqaRetrievalself-reported49.266
- cos_sim_accuracy on MTEB Cmnlivalidation set self-reported86.085
- cos_sim_ap on MTEB Cmnlivalidation set self-reported92.644
- cos_sim_f1 on MTEB Cmnlivalidation set self-reported86.900
- cos_sim_precision on MTEB Cmnlivalidation set self-reported84.114
- cos_sim_recall on MTEB Cmnlivalidation set self-reported89.876
- dot_accuracy on MTEB Cmnlivalidation set self-reported72.664
- dot_ap on MTEB Cmnlivalidation set self-reported81.053
- dot_f1 on MTEB Cmnlivalidation set self-reported75.199
- dot_precision on MTEB Cmnlivalidation set self-reported67.491
- dot_recall on MTEB Cmnlivalidation set self-reported84.896
- euclidean_accuracy on MTEB Cmnlivalidation set self-reported85.520
- euclidean_ap on MTEB Cmnlivalidation set self-reported91.906
- euclidean_f1 on MTEB Cmnlivalidation set self-reported86.264
- euclidean_precision on MTEB Cmnlivalidation set self-reported82.207
- euclidean_recall on MTEB Cmnlivalidation set self-reported90.741
- manhattan_accuracy on MTEB Cmnlivalidation set self-reported85.484
- manhattan_ap on MTEB Cmnlivalidation set self-reported91.897
- manhattan_f1 on MTEB Cmnlivalidation set self-reported86.204
- manhattan_precision on MTEB Cmnlivalidation set self-reported84.325
- manhattan_recall on MTEB Cmnlivalidation set self-reported88.169
- max_accuracy on MTEB Cmnlivalidation set self-reported86.085
- max_ap on MTEB Cmnlivalidation set self-reported92.644
- max_f1 on MTEB Cmnlivalidation set self-reported86.900